the jupyter notebook as an interface to a larger code base
Although the Jupyter Notebook makes a great code sandbox, and while there are many eloquent notebooks online detailing, say, how to analyze a specific dataset in a specific way, it’s generally not terribly efficient to put your entire project inside a single notebook. The reason is simply that keeping everything inside a lone, linearly structured file can make it challenging to keep track of and manipulate all the moving pieces. It would be much easier to find a particular statistical function you’ve written, for instance, by looking in a module called my_stats.py inside a directory called analysis than by scrolling through an extensive webpage. In addition, the latter organizational strategy is much more standard, which can be a good thing in software design.
This doesn’t mean, however, that Jupyter Notebooks are only useful as a sandbox and for tutorials and have no place in your “production-level” code. As the title suggests, I’ve found that the Jupyter Notebook also serves as a very practical interface or control panel to your main code base. That is, instead of executing your code by running a script from the command line or from within an IDE (by pressing F5 or clicking on a green run button, for example), you can call a function you’ve written from within a Jupyter Notebook code cell. For example, in the excerpt below I’ve called the function lif_neuron_white_noise_input from within figures/model_neuron_demos.py. This is a function that simulates the response of a model neuron to a random electrical stimulus (see the Wikipedia page for more details if you’re interested).
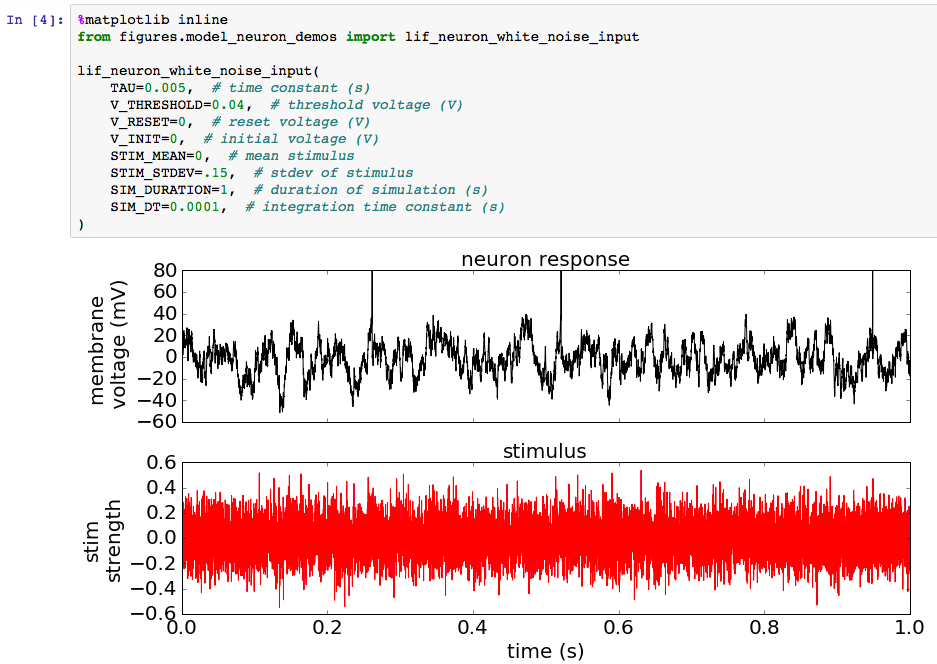
Calling a function you’ve written from inside a notebook cell comes with a number of advantages. First, as detailed in the last post, Jupyter Notebooks allow you to cleanly display textual, tabular, and graphical output without any fuss over graphical backends or configurations or formatting. Further, you can surround your output with markdown (text with a shorthand syntax for elegant formatting), equations, images, hyperlinks, etc. to introduce background or explain what your code is doing. Also nice is that since the output is saved in the file, a notebook provides a clean log of the times you’ve executed a portion of your code base along with the outputs that resulted.
A bit more subtly, having a control panel for your main code base also forces you to ask what the natural inputs to each analysis/simulation function are, i.e., what aspects or parameters of your code are most useful to keep explicitly modifiable. In the example above, for instance, you can specify the threshold and time constant of the simulated neuron, the amplitude of the stimulus, etc., and these inputs are instantiated simply as arguments of the lif_neuron_white_noise_input function call inside the notebook. Using function arguments to specify your inputs, as is completely natural in a Jupyter Notebook, is far simpler than developing a complicated graphical user interface (GUI) and allows for a large amount of on-the-fly flexibility (for example, in Python you could also pass in a quick code snippet like a list comprehension as an argument), while retaining a clean visual experience. Also nice about this approach is that the parameters of the analysis/simulation are kept in a separate file (the Jupyter Notebook) from the code for the function itself. Thus, once a function is written and tested, you can essentially leave its code alone, which is both good practice in software design and makes it easier to manage your project with a standard version control system.
Building off the last point, I’ve often found myself needing to share my work with readers who are primarily interested in the parameters and results of an analysis/simulation and only secondarily (or not at all) in the nuts and bolts of the actual code. Fortunately, interfacing with a code base in the way I’ve been describing makes your work ideally suited to sharing with this sort of reader. For if someone were to read our notebook in the example above, she would see the name of the function we’d called, the inputs/arguments to that function, the graphical result, and perhaps an accompanying description of the background or mathematics. She could therefore immediately view the choices we’d made for each decision we’d deemed important for the simulation, and she could examine the results immediately below it without being overwhelmed by every line of code that went into it. If she wanted to see how the results changed with different function arguments, she’d have only to edit that notebook cell and run it, without any fuss over digging through a more complicated code base. Of course, if she wanted to see the underlying code that would be simple too, since the import statements specify exactly where each the code for each function lives.
Speaking of sharing notebooks, Jupyter Notebooks happen to render on GitHub, so sharing a static notebook with someone is as simple as pushing the notebook to an online repository and sharing the link.
Finally, one of the most useful aspects of using Jupyter Notebooks to interface with a larger code base I’ve encountered is that you can easily use them to run code on a remote computer. This results directly from the Jupyter application running out of a web server and using a web browser as its user interface. A notebook server can be started on one computer and accessed by another simply by navigating to a specific URL, which will bring up the notebook application. Then when you use the browser on your local computer to execute a code cell, the code gets run on the remote computer but the output gets rendered in the browser of your local computer in exactly the same way as if there were no remote connection at all.
In fact, when I made the example notebook above, I wrote and executed the simulation and viewed the graphical output purely from home on my personal laptop, but the code itself ran on a computer in my lab. Thus, the web browser acts like a portal into a code base on either your local or a remote machine which cleanly handles all aspects of the display. Compared to running remote code by screen-sharing, which is notoriously buggy, laggy, operating-system-dependent, and transmits way more graphical data than you typically actually need, or by sshing into a remote computer, executing a script from the command line, and then either attempting to pipe graphical output through the ssh tunnel (which is not that easy and certainly not that flexible) or saving it to the remote disk and syncing it via Dropbox or the like to your local computer to subsequently open it, running remote code using Jupyter is marvelously simple. You can even have interactive outputs rendered on your local browser that resulted from code running on a remote computer.
If you want to run a Jupyter Notebook server remotely yourself, check out this post, which explains the process in some detail.
In all, I’ve found that using Jupyter Notebooks as the interface with my primary code base has solved a lot of headache-inducing problems and has greatly simplified my workflow, and I hope that I’ve managed here to illuminate some of the reasons for using Jupyter in this context.